Relearning R

A hodgepodge of notes for learning R for my reference, segmented so it is easy to read.
–
Visualisations
ggplot(data = <DATA>) +
<GEOM_FUNCTION>(
mapping = aes(<MAPPINGS>),
stat = <STAT>,
position = <POSITION>
) +
<COORDINATE_FUNCTION> +
<FACET_FUNCTION>
- Limit axes:
+ coord_cartesian(ylim = c(0, 50))- Use xlim for x-axis
- use
facets: subplots that display subsets of data (categorical variables)
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy)) +
facet_wrap(~ class, nrow = 2)
-
also can use
facet_grid()to plot combination of two variables:ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy)) + facet_grid(drv ~ cyl) -
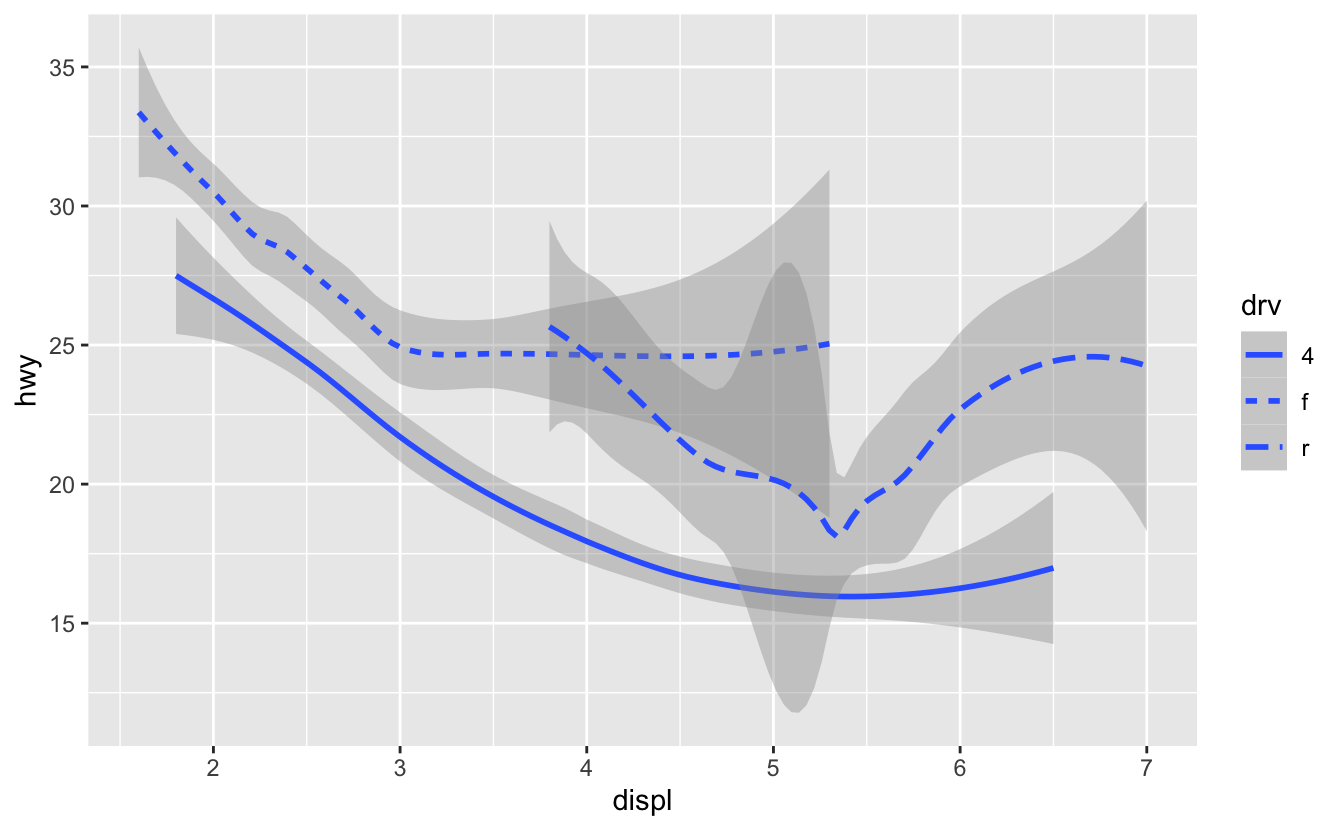
geom_pointvsgeom_smooth; which object to map plots; every geom function takesmappingas an argument; instead oflinetypebelow, can also usegroupto show different types of categories with same linetypeggplot(data = mpg) + geom_smooth(mapping = aes(x = displ, y = hwy, linetype = drv))
-
repetition isn’t necessary:
ggplot(data = mpg, mapping = aes(x = displ, y = hwy)) + geom_point(mapping = aes(color = class)) + geom_smooth(data = filter(mpg, class == "subcompact"), se = FALSE) -
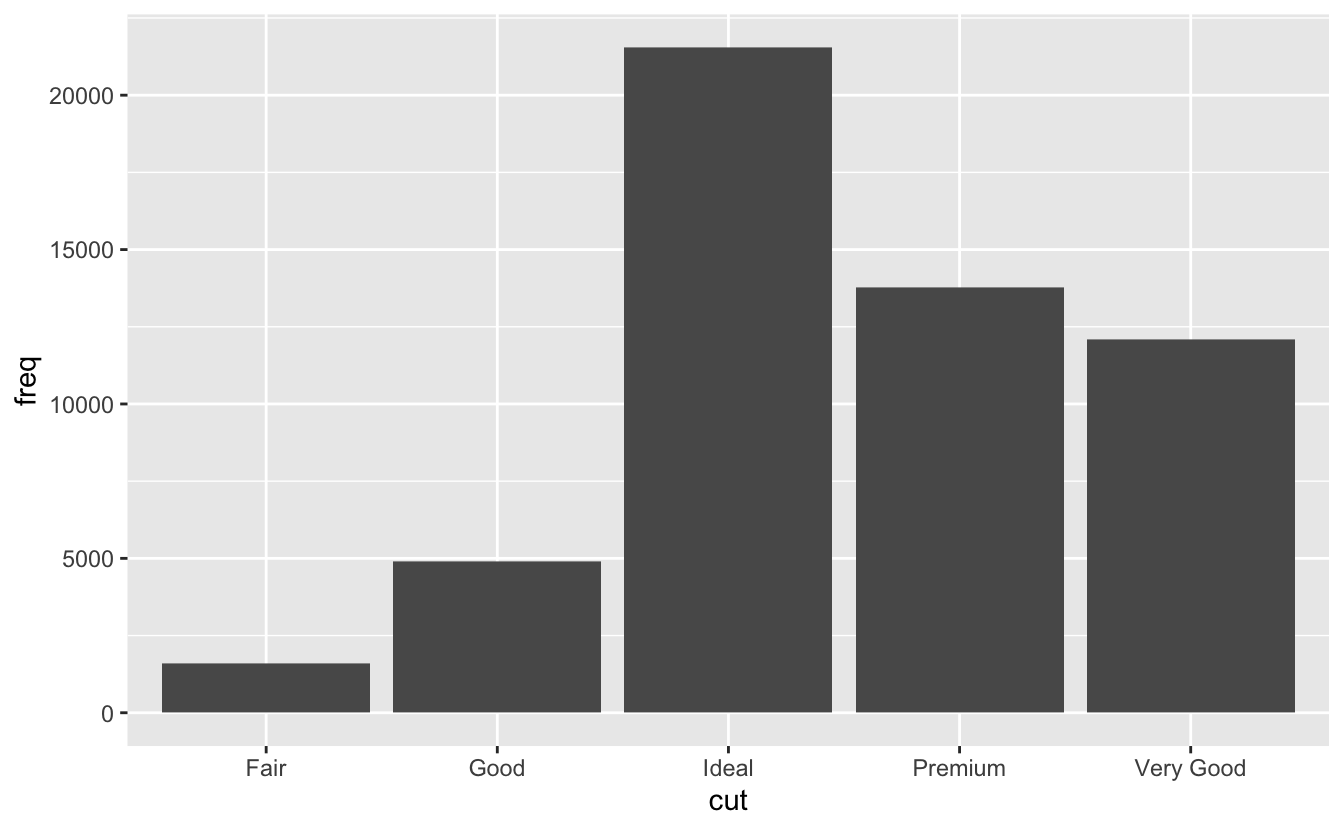
changing order of bar chart for categorical mappings (so doesn’t order based on increasing frequency):
demo <- tribble( ~cut, ~freq, "Fair", 1610, "Good", 4906, "Very Good", 12082, "Premium", 13791, "Ideal", 21551 ) ggplot(data = demo) + geom_bar(mapping = aes(x = cut, y = freq), stat = "identity" #use y = stat(prop) for proportion
-
lineplots (w stats)/ boxplots:
ggplot(data = diamonds) + stat_summary( mapping = aes(x = cut, y = depth), fun.min = min, fun.max = max, fun = median ) ggplot(data = mpg, mapping = aes(x = class, y = hwy)) + geom_boxplot() #add + coord_flip() to make it horizontal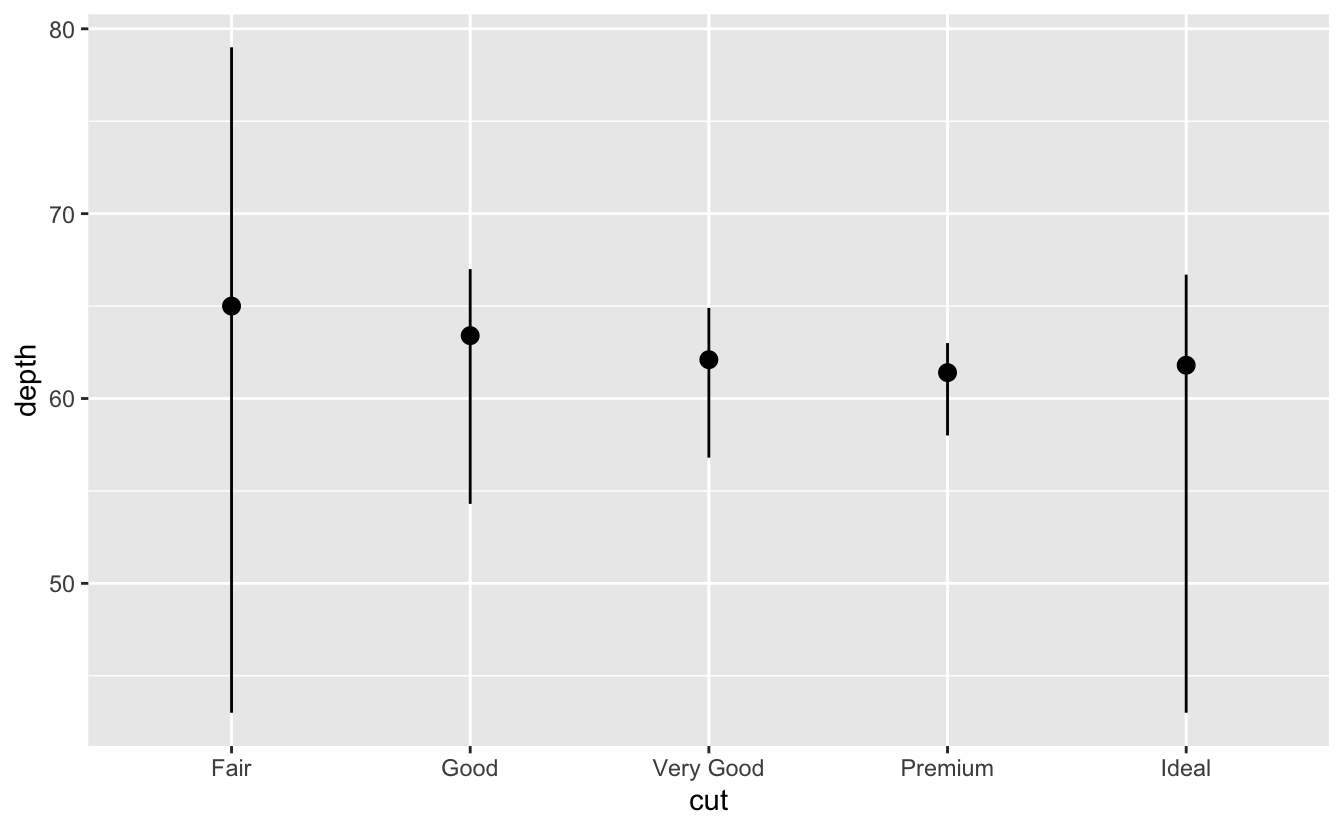
-
Use
reorderto reorder boxplots (FUNis function to use)ggplot(data = mpg) + geom_boxplot(mapping = aes(x = reorder(class, hwy, FUN = median), y = hwy)) -
can use
fill(also for categorical) to fill whole bars, or normalcolorfor outlines -
+ coord_flip()flips the axis -
Different boxplot width based on # of points in each bin:
ggplot(data = smaller, mapping = aes(x = carat, y = price)) + geom_boxplot(mapping = aes(group = cut_number(carat, 20))) -
position = "identity"will place each object exactly where it falls in the context of the graph. This is not very useful for bars, because it overlaps them. To see that overlapping we either need to make the bars slightly transparent by settingalphato a small value, or completely transparent by settingfill = NA; basically doesn’t stack like fill, but overlaps on top of each other to see original value compared to y -
position = "fill"works like stacking, but makes each set of stacked bars the same height. This makes it easier to compare proportions across groups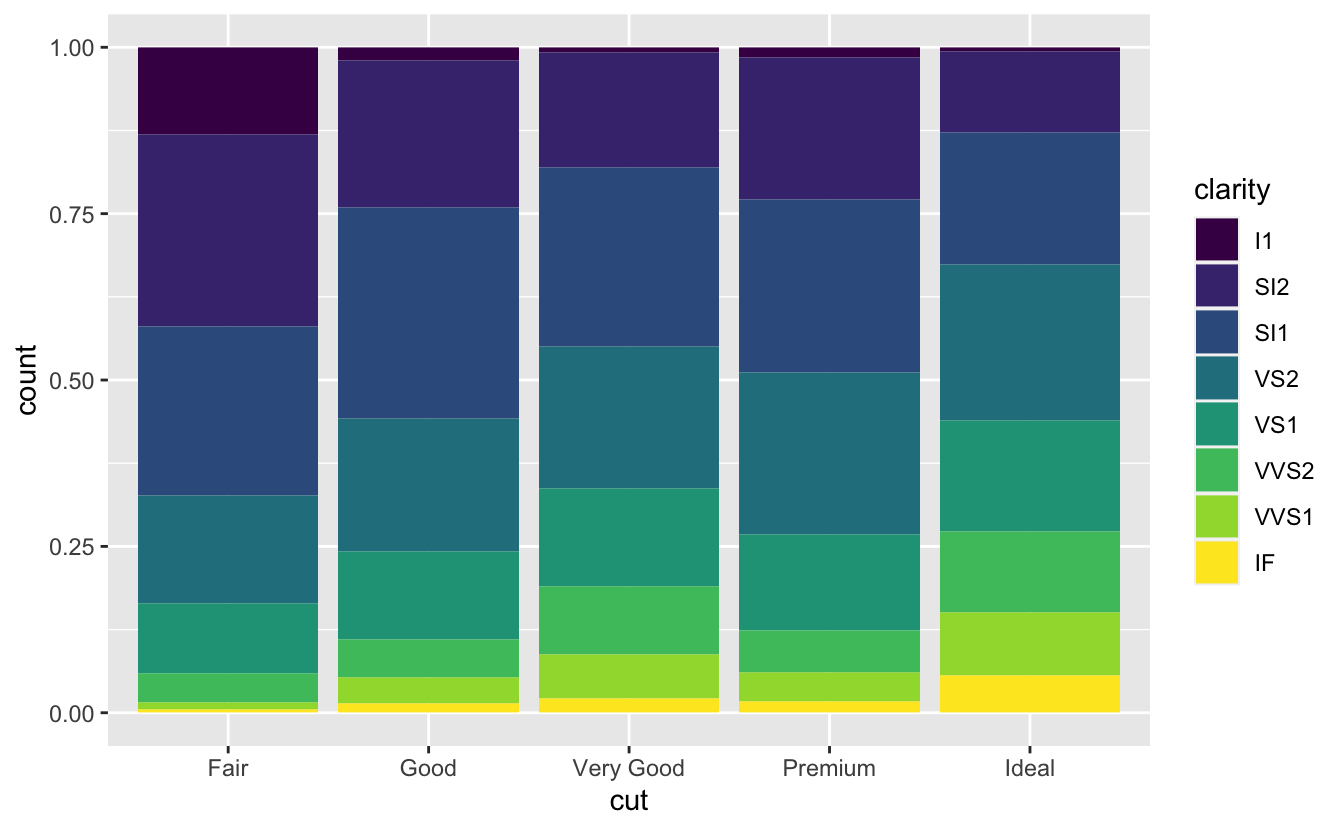
-
position = "dodge"places overlapping objects directly beside one another. This makes it easier to compare individual values; below image shows this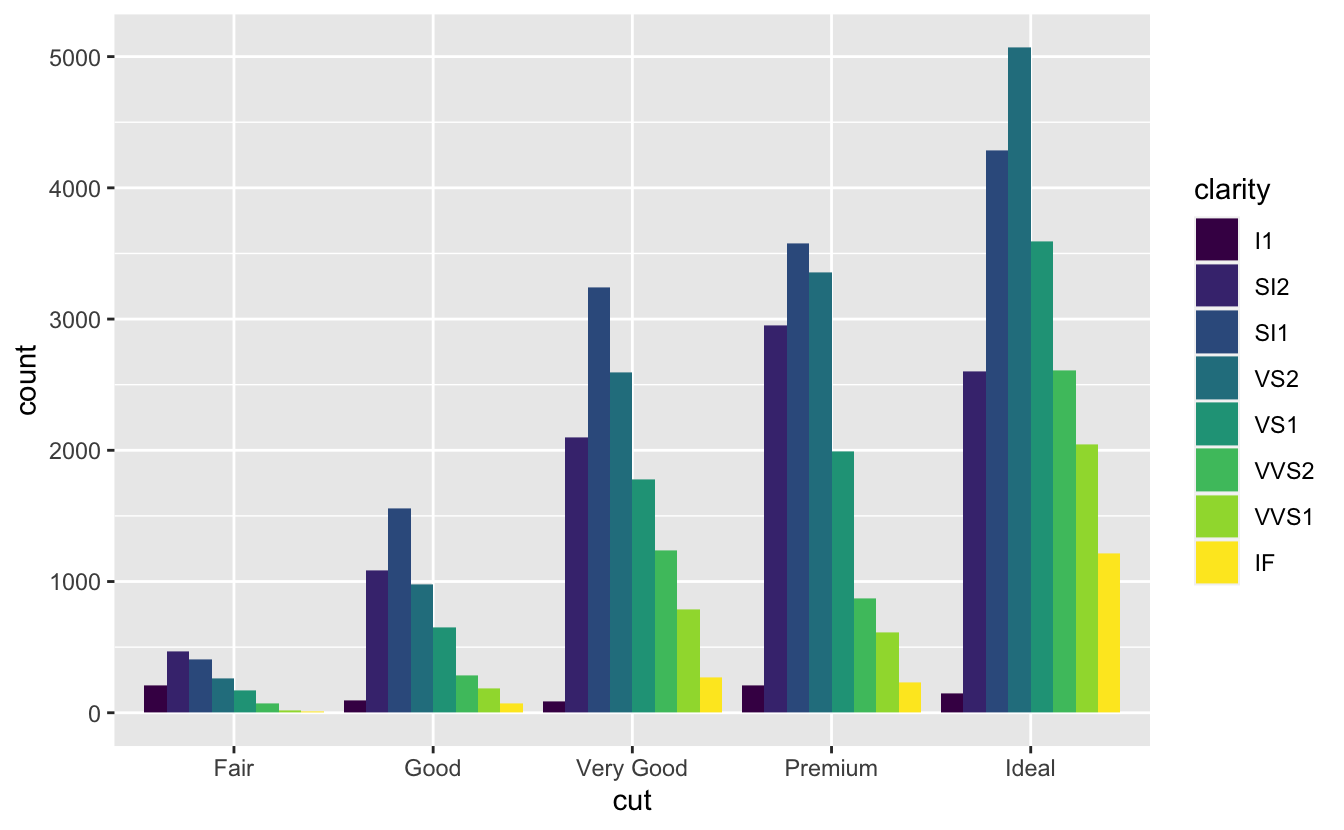
-
use position =
jitterfor scatterplot when points over plot with each other; + geom_jitter() -
can use
bar + coord_polar()to make bar charts into pie charts -
explore labs for tags/titles/etc
-
Instead of using
geom_histogramusegeom_freqpolyfor categories of data:ggplot(diamonds, aes(price, colour = cut)) + geom_freqpoly(binwidth = 500)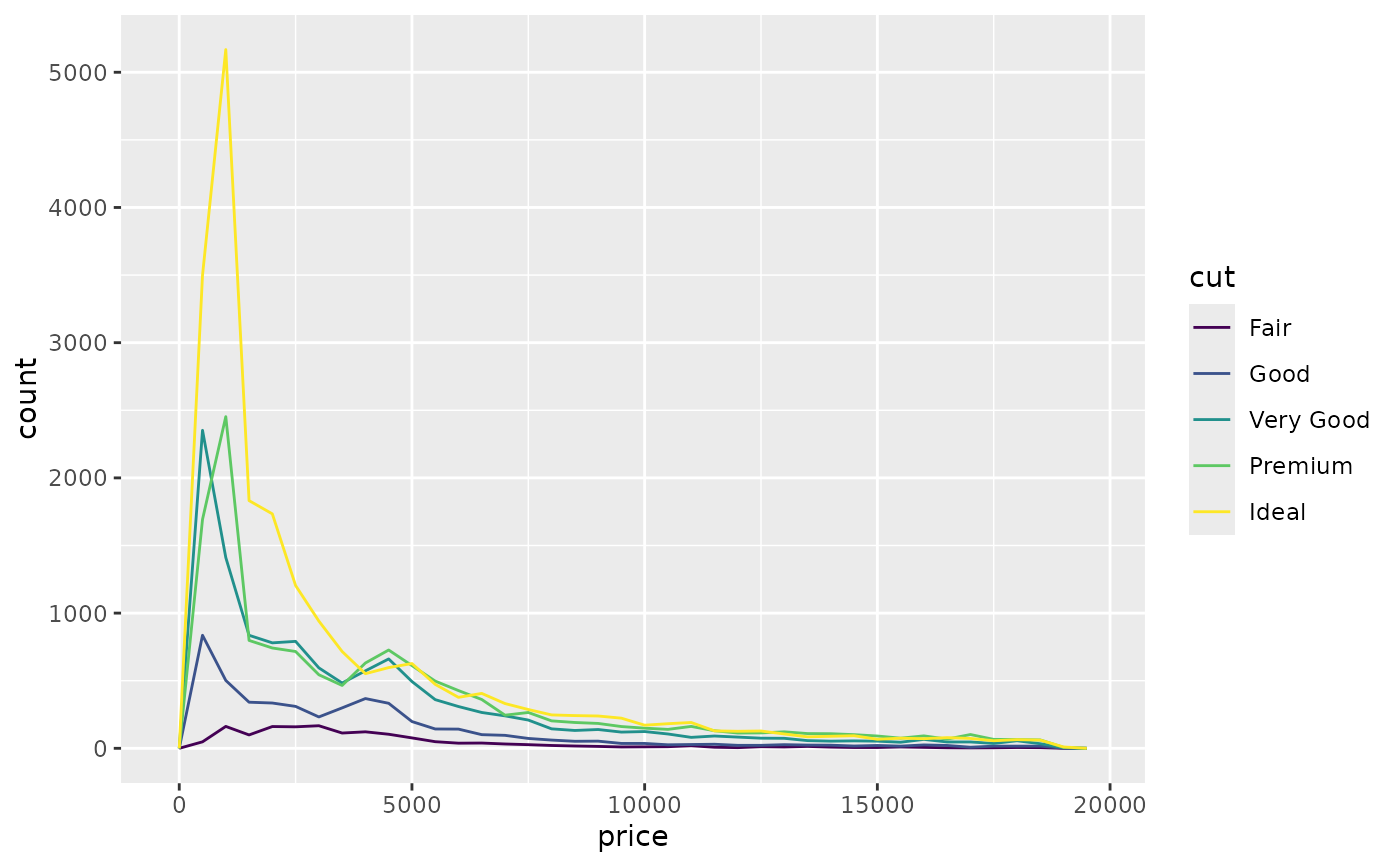
–
The Basics
-
Do not use
=, instead doALT+-to create<-to assign variables -
Use snake case for styling convention (underscore to separate lowercase):
example:
i_use_snake_case -
Use
CTRL+UP_ARROWto look at all commands executed in terminal (fast-ease to re-do past commands) -
Variables are case-sensitive – typos will happen
-
When shifting through different functions using
TABautofill, pressF1to get more detailed documentation -
ALT+SHIFT+Kto access all keyboard shortcuts -
Press
Ctrl + Shift + F10to restart RStudio -
Press
Ctrl + Shift + Sto rerun the current script -
dplyroverwrites base functions of R – to access original, use:stats::function -
Easy access of functions / objects using
::
- one example:
nycflights13::flightsto access dataframe
-
To print and save assignments, wrap in parentheses:
- example:
(dec25 <- filter(flights, month == 12, day == 25))
- example:
-
When using
==to check for boolean conditions, usenearinstead:sqrt(2) ^ 2 == 2 #> [1] FALSE 1 / 49 * 49 == 1 #> [1] FALSE -- near(sqrt(2) ^ 2, 2) #> [1] TRUE near(1 / 49 * 49, 1) #> [1] TRUE -
Ctrl + Shift + P to resend the complete block that was recently used in console– useful when editing a block constantly.
Data Transformation
-
tibblesare dataframes but tweaked to work undertidyverse -
vectors can be named (used for subsetting):
-
c(x = 1, y = 2, z = 4) #or set_names(1:3, c("a", "b", "c")) #> a b c #> 1 2 3
-
-
subsetting starts at index 1:
-
x <- c("one", "two", "three", "four", "five") x[c(3, 2, 5)] #> [1] "three" "two" "five" -
negative values drop element, and mixing negative and positive do not work:
x[c(-1, -3, -5)] #> [1] "two" "four" x[c(1, -1)] #> Error in x[c(1, -1)]: only 0's may be mixed with negative subscripts -
subsetting in lists with [] returns smaller list, [[]] gets element:
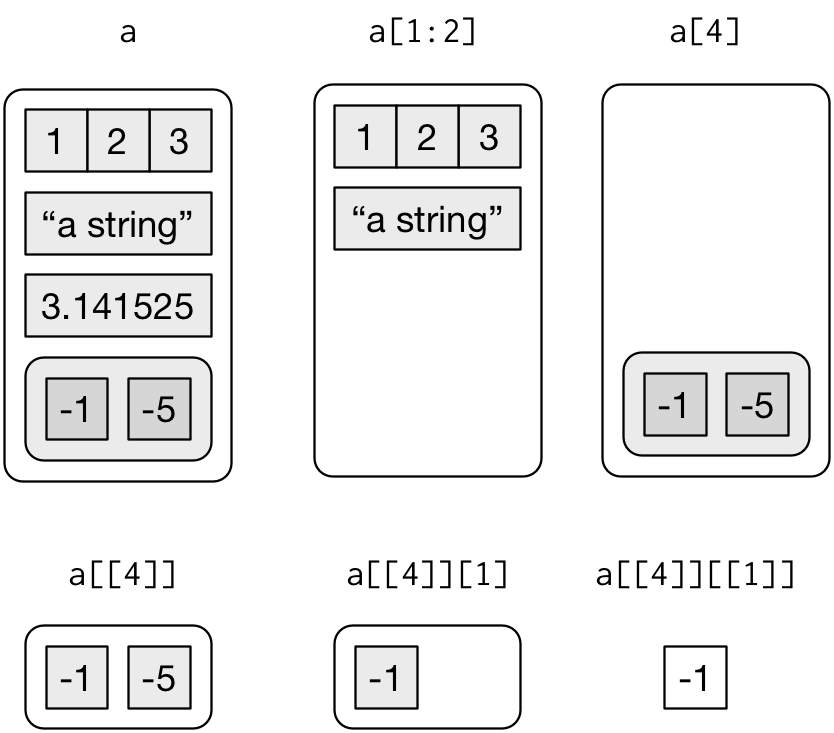
-
-
&= “and”,|= “or”,!= “not”-
Boolean operations in picture:
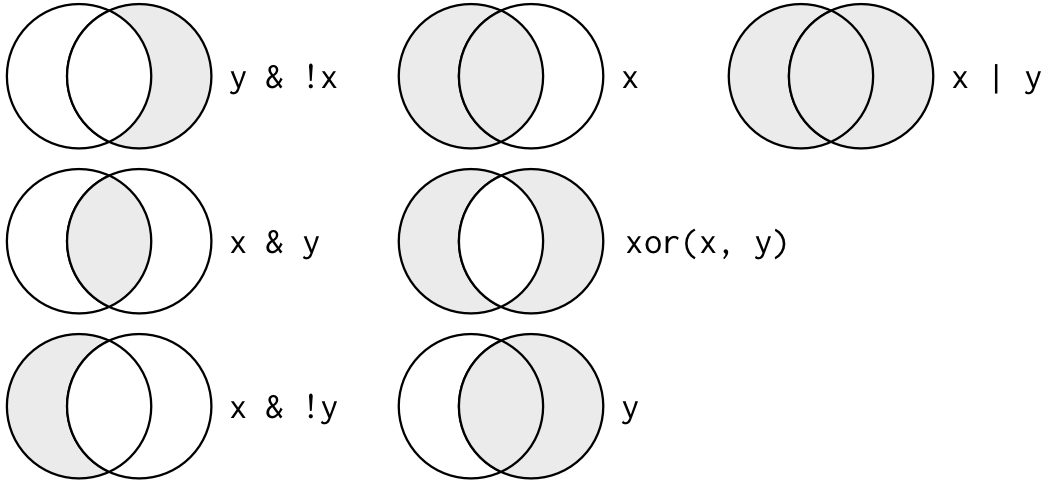
-
-
the following abbreviations for types of variables:
int: integer,dbl: doubles/real numberschr: character vectors, or stringsdttm: date-times (date+time)lgl: logical (booleans), vectors with onlyTRUEorFALSEfctr: factors (used to represent categorical variables with fixed possible values)
–
-
the basics of
dplyr, in which the first argument is adf:-
Pick observations by their values filter().
-
example:
filter(flights, !(arr_delay > 120 | dep_delay > 120)) filter(flights, arr_delay <= 120, dep_delay <= 120) #Demorgans law: !(x & y) is the same as !x | !y, #and !(x | y) is the same as !x & !y -
can use
between(x, left, right)instead of boolean condition statements: ` x >= left & x <= right`
-
-
Reorder the rows: arrange()
- selects rows in ascending order, can use
desc()to re-order in descending order:arrange(flights, desc(dep_delay))
- selects rows in ascending order, can use
-
Pick variables by their names:
select().- Can select columns between two variables:
select(flights, year:day) - Can select columns not between two variables (inclusive):
select(flights, -(year:day)) - Some helper functions, see
?selectfor more:starts_with("abc")ends_with("xyz")contains("ijk")everything()pulls all the other variables to re-ordernum_range("x", 1:3)matches variablesx1tox3
- Use
renameto keep all variables not mentioned (as select drops it)rename(flights, tail_num = tailnum)
- Can select columns between two variables:
-
Create new variables with functions of existing variables: mutate().
-
similar to
apply()from python; can also refer to columns that you’ve created in the same line; ONLY TEMPORARY unless stored -
function must be vectorized: it must take a vector of values as input, return a vector with the same number of values as output.
-
adds new variable to end of dataset; use
View(df)to see all columns -
Example:
flights_sml <- select(flights, year:day, ends_with("delay"), distance, air_time ) mutate(flights_sml, gain = dep_delay - arr_delay, speed = distance / air_time * 60 ) #> # A tibble: 336,776 x 9 #> year month day dep_delay arr_delay distance air_time gain speed #> <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 2013 1 1 2 11 1400 227 -9 370. -
if you only want to keep the new variables, use
transmute() -
%/%= integer division, dividing and then rounding down to nearest integer -
%%= mod, or just remainder -
Other functions:
lead(),lag()(refers to the number that leads them or is behind them),cumsum(),cumprod(),cummin(),cummax(),cummean() -
Also ranking functions:
min_rank(),row_number(),dense_rank(),percent_rank(),cume_dist(),ntile()transmute(flights, dep_time, hour = dep_time %/% 100, minute = dep_time %% 100 ) #> # A tibble: 336,776 x 3 #> dep_time hour minute #> <int> <dbl> <dbl> #> 1 517 5 17 #> 2 533 5 33 #> 3 542 5 42 #> 4 544 5 44 #> 5 554 5 54 #> 6 554 5 54 #> # … with 336,770 more rows
-
-
Collapse many values down to a single summary: summarise().
- use in tandem with
group_byto compute unit of analysis from complete dataset to individual groups. count = n()as a sub function to calculate # of data pointssum(!is.na(x))as a sub function to calculate # of missing valuesn_distinct(x)for distinct values
- or can calculate statistics of overall data-frame like mean:
summarise(flights, delay = mean(dep_delay, na.rm = TRUE))
- use in tandem with
-
And then can operate by groups: group_by().
- use
ungroup()to ungroup back to one layer - also use filters and mutations on groups
- use
-
**Piping: **
%>%or read as “then,” which combines all transformations into one code block and it’s easier to read:- sequential:
x %>% f(y)turns intof(x, y), andx %>% f(y) %>% g(z)turns intog(f(x, y), z)and so on
by_dest <- group_by(flights, dest) delay <- summarise(by_dest, count = n(), dist = mean(distance, na.rm = TRUE), delay = mean(arr_delay, na.rm = TRUE) ) #> `summarise()` ungrouping output (override with `.groups` argument) delay <- filter(delay, count > 20, dest != "HNL") ## VS. Piping way delays <- flights %>% group_by(dest) %>% summarise( count = n(), dist = mean(distance, na.rm = TRUE), delay = mean(arr_delay, na.rm = TRUE) ) %>% filter(count > 20, dest != "HNL") - sequential:
-
sum of one variable (total # of miles a certain plane flew), where
wtrefers to weight of the variable being counted.- think of this as a sum of a variable based on another variable
not_cancelled %>% count(tailnum, wt = distance) #> # A tibble: 4,037 x 2 #> tailnum n #> <chr> <dbl> #> 1 D942DN 3418 #> 2 N0EGMQ 239143 #> 3 N10156 109664 #> 4 N102UW 25722 #> 5 N103US 24619 #> 6 N104UW 24616 #> # … with 4,031 more rows
-
Handling Missing Values
-
NArepresents unknown values, and any operations involving will also be unknown-
NA == NAwill result inNAgiven this same premise -
use
is.na(x)to determine if value is missing -
can be persevered when filtering:
filter(df, is.na(x) | x > 1) -
When working with
arrange(), missing values will always be sorted at the end, use indicators to have them at the beginning:arrange(flights, desc(is.na(dep_time)), dep_time)
-
–
Data Exploration/Transformation
Questions to Explore:
- Which values are the most common? Why?
- Which values are rare? Why? Does that match your expectations?
- Can you see any unusual patterns? What might explain them?
-
Change certain values of column through boolean condition:
diamonds2 <- diamonds %>% mutate(y = ifelse(y < 3 | y > 20, NA, y))-
Change
y: if it meets condition thenNA, elsey; -
Look into
case_when()to create a new variable that does complex combinations of existing variables
-
-
Two ways to visualize covariation with categorical variables:
-
diamonds %>% count(color, cut) %>% ggplot(mapping = aes(x = color, y = cut)) + geom_tile(mapping = aes(fill = n)) -
ggplot(data = diamonds) + geom_count(mapping = aes(x = cut, y = color))
Using map instead of for loops:
map()makes a list.map_lgl()makes a logical vector.map_int()makes an integer vector.map_dbl()makes a double vector.map_chr()makes a character vector.
–
Tibbles:
-
Convert df to tibbles:
as_tibble(df) -
Convert vector to tibbles:
tibble( x = 1:5, y = 1, z = x ^ 2 + y)- variables x,y,z as functions
-
Use ` (backticks) to use non-synaptic names
-
tb <- tibble( `:)` = "smile", ` ` = "space", `2000` = "number")
-
-
Tribbles, create tibbles using tables directly:
tribble(
~x, ~y, ~z,
#--|--|----
"a", 2, 3.6,
"b", 1, 8.5
)
-
Different with printing and subsetting (shows only the first 10 rows, and all the columns that fit on screen, each column reports its type, a nice feature borrowed from str
-
[[can extract by name or position;$only extracts by name but is a little less typing- tibbles can’t do partial matching
-
Can convert back to df:
class(as.data.frame(tb))#tb is tibbles
–
Importing and Reading Data (using readrof tidyverse):
-
read_csv()reads comma delimited files or inline csv files,read_csv2()reads semicolon separated files,read_tsv()reads tab delimited files, andread_delim()reads in files with any delimiter.a.)
na: this specifies the value (or values) that are used to represent missing values in your fileb.) the data might not have column names. you can use
col_names = FALSEto tell read_csv() not to treat the first row as headings, and instead label them sequentially fromX1toXn; alternatively you can passcol_namesa character vector which will be used as the column namesc.) sometimes there are a few lines of metadata at the top of the file: use
skip = nto skip the first n lines; usecomment = "#"to drop all lines that start with (e.g.) # -
read_fwf()reads fixed width files. You can specify fields either by their widths withfwf_widths()or their position withfwf_positions()a)
read_table()reads a common variation of fixed width files where columns are separated by white space. -
read_log()reads Apache style log files -
First argument is the path
-
Compared to base R,
tidyversecreates tibbles and doesn’t convert character vectors to factors
–
Parsing Data:
-
Like all functions in the tidyverse, the parse_*() functions are uniform: the first argument is a character vector to parse, and the na argument specifies which strings should be treated as missing
parse_logical()andparse_integer()parse logicals and integers respectively. There’s basically nothing that can go wrong with these parsers so I won’t describe them here furtherparse_double()is a strict numeric parser, and parse_number() is a flexible numeric parser. These are more complicated than you might expect because different parts of the world write numbers in different waysparse_character()seems so simple that it shouldn’t be necessary. But one complication makes it quite important: character encodingsparse_factor()create factors, the data structure that R uses to represent categorical variables with fixed and known valuesparse_datetime(),parse_date(), andparse_time()allow you to parse various date & time specifications. These are the most complicated because there are so many different ways of writing dates- readr has the notion of a
“locale”, an object that specifies parsing options that differ from place to place. When parsing numbers, the most important option is the character you use for the decimal mark. You can override the default value of parse_number()addresses the second problem: it ignores non-numeric characters before and after the number- you need to specify the encoding in parse_character(): parse_character(x1, locale = locale(encoding = “Latin1”)); use guess_encoding() to help you figure it out
- readr also comes with two useful functions for writing data back to disk:
write_csv()andwrite_tsv() - Every
parse_xyz()function has a correspondingcol_xyz()function. You use parse_xyz() when the data is in a character vector in R already; you use col_xyz() when you want to tell readr how to load the data.
Example:
challenge <- read_csv( readr_example("challenge.csv"), col_types = cols( x = col_double(), y = col_date() )) #Sometimes it’s easier to diagnose problems if you just read in all the columns as character vectors: challenge2 <- read_csv(readr_example("challenge.csv"), col_types = cols(.default = col_character()) )Custom Date-Time Format Parsing:
-
Year:
- %Y (4 digits).
- %y (2 digits); 00-69 -> 2000-2069, 70-99 -> 1970-1999.
-
Month:
- %m (2 digits).
- %b (abbreviated name, like “Jan”).
- %B (full name, “January”).
-
Day:
- %d (2 digits).
- %e (optional leading space).
-
Time:
- %H 0-23 hour.
- %M minutes; %S integer seconds%OS real seconds.
- %I 0-12, must be used with %p.
- %p AM/PM indicator.
- %Z Time zone (as name, e.g. America/Chicago). Beware of abbreviations: if you’re American, note that “EST” is a Canadian time zone that does not have daylight savings time. It is not Eastern Standard Time.
- %z (as offset from UTC, e.g. +0800).
- %H 0-23 hour.
-
Non-digits:
- %. skips one non-digit character.
- %* skips any number of non-digits.
–
Tidy Data:
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.
There’s a specific advantage to placing variables in columns because it allows R’s vectorised nature to shine. As you learned in mutate and summary functions, most built-in R functions work with vectors of values. That makes transforming tidy data feel particularly natural.
-
pivot_longer: change observations into columnstable4a #> # A tibble: 3 x 3 #> country `1999` `2000` #> * <chr> <int> <int> #> 1 Afghanistan 745 2666 #> 2 Brazil 37737 80488 #> 3 China 212258 213766 table4a %>% pivot_longer(c(`1999`, `2000`), names_to = "year", values_to = "cases") #> # A tibble: 6 x 3 #> country year cases #> <chr> <chr> <int> #> 1 Afghanistan 1999 745 #> 2 Afghanistan 2000 2666 #> 3 Brazil 1999 37737 #> 4 Brazil 2000 80488 #> 5 China 1999 212258 #> 6 China 2000 213766 -
Similar to
pivot_wider:
–
Memory:
pryr::object_size() gives the memory occupied by all of its arguments.
- If two dataframes share the same columns from mutations, etc. then they have the same memory; if one value of one dataframe is changed, then memory is increased since column can no longe rbe shared
–
When to not pipe (%>%)
The pipe is a powerful tool, but it’s not the only tool at your disposal, and it doesn’t solve every problem! Pipes are most useful for rewriting a fairly short linear sequence of operations. I think you should reach for another tool when:
- Your pipes are longer than (say) ten steps. In that case, create intermediate objects with meaningful names. That will make debugging easier, because you can more easily check the intermediate results, and it makes it easier to understand your code, because the variable names can help communicate intent.
- You have multiple inputs or outputs. If there isn’t one primary object being transformed, but two or more objects being combined together, don’t use the pipe.
- You are starting to think about a directed graph with a complex dependency structure. Pipes are fundamentally linear and expressing complex relationships with them will typically yield confusing code.
–
Coercion:
- converting one variable type to another
-
Explicit coercion happens when you call a function like
as.logical(),as.integer(),as.double(), oras.character()- should always try to upstream the change first so the vector isn’t read wrong in the first place (tweak readr col_types)
-
Implicit coercion happens when you use a vector in a specific context that expects a certain type of vector. For example, when you use a logical vector with a numeric summary function, or when you use a double vector where an integer vector is expected.
-
Most complex variables of a vector wins for the type of the list:
typeof(c(TRUE, 1L)) #> [1] "integer" typeof(c(1L, 1.5)) #> [1] "double" typeof(c(1.5, "a")) #> [1] "character" -
Use the following instead of general
is_vector:lgl int dbl chr list is_logical()x is_integer()x is_double()x is_numeric()x x is_character()x is_atomic()x x x x is_list()x is_vector()x x x x x
–
For more resources:
- Notes taken from an online book here