Python Integration in RStudio?
It’s finally here! Well, it has been here actually. The reticulate package has provided tools for interoperability between Python and R; it allows:
- Calling Python from R in a variety of ways including R Markdown, sourcing Python scripts, importing Python modules, and using Python interactively within an R session.
- Translation between R and Python objects (for example, between R and Pandas data frames, or between R-matrices and NumPy arrays).
- Flexible binding to different versions of Python including virtual environments and Conda environments.
So What Changed?
Through the new update for RStudio 1.4, the Python editing experience has been improved even further:
- The default Python interpreter to be used by RStudio /
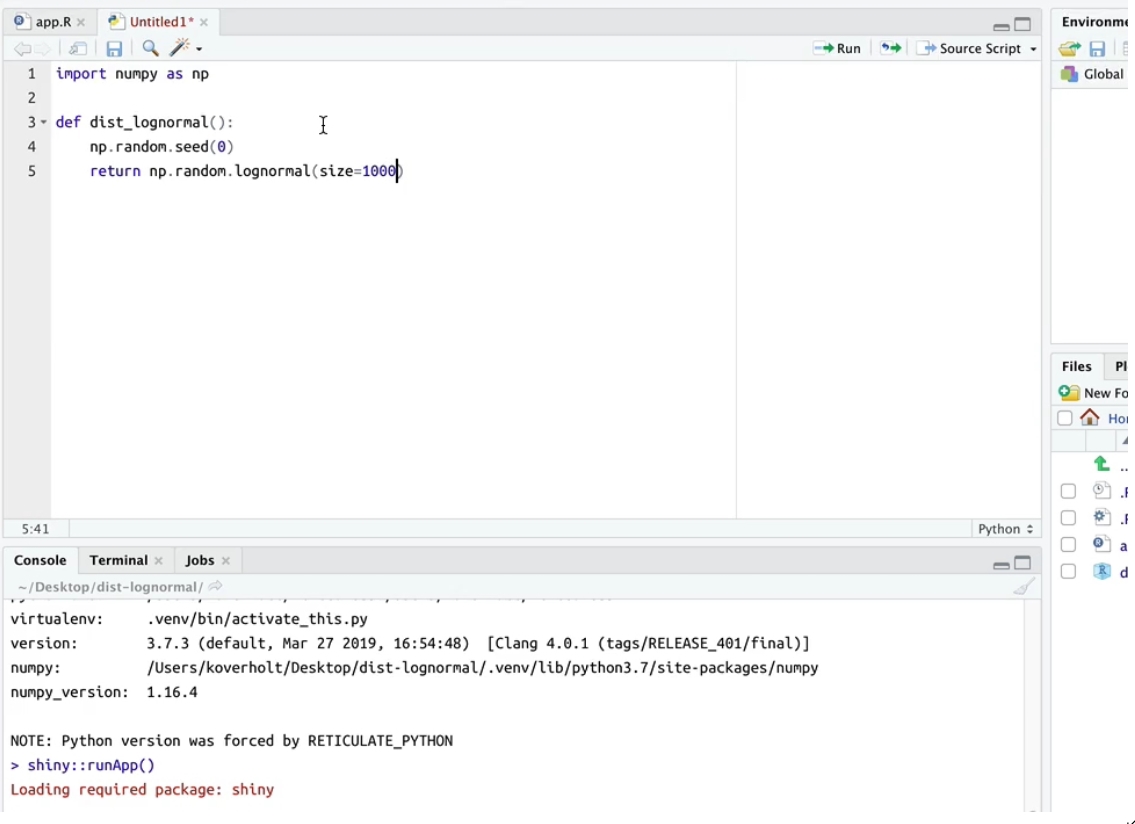
reticulatecan now be customized in the Global Options pane. - The Environment pane now displays a summary of Python objects available in the main module when the
reticulateREPL is active. - Python objects can now be viewed and explored within the RStudio data viewer and object explorer.
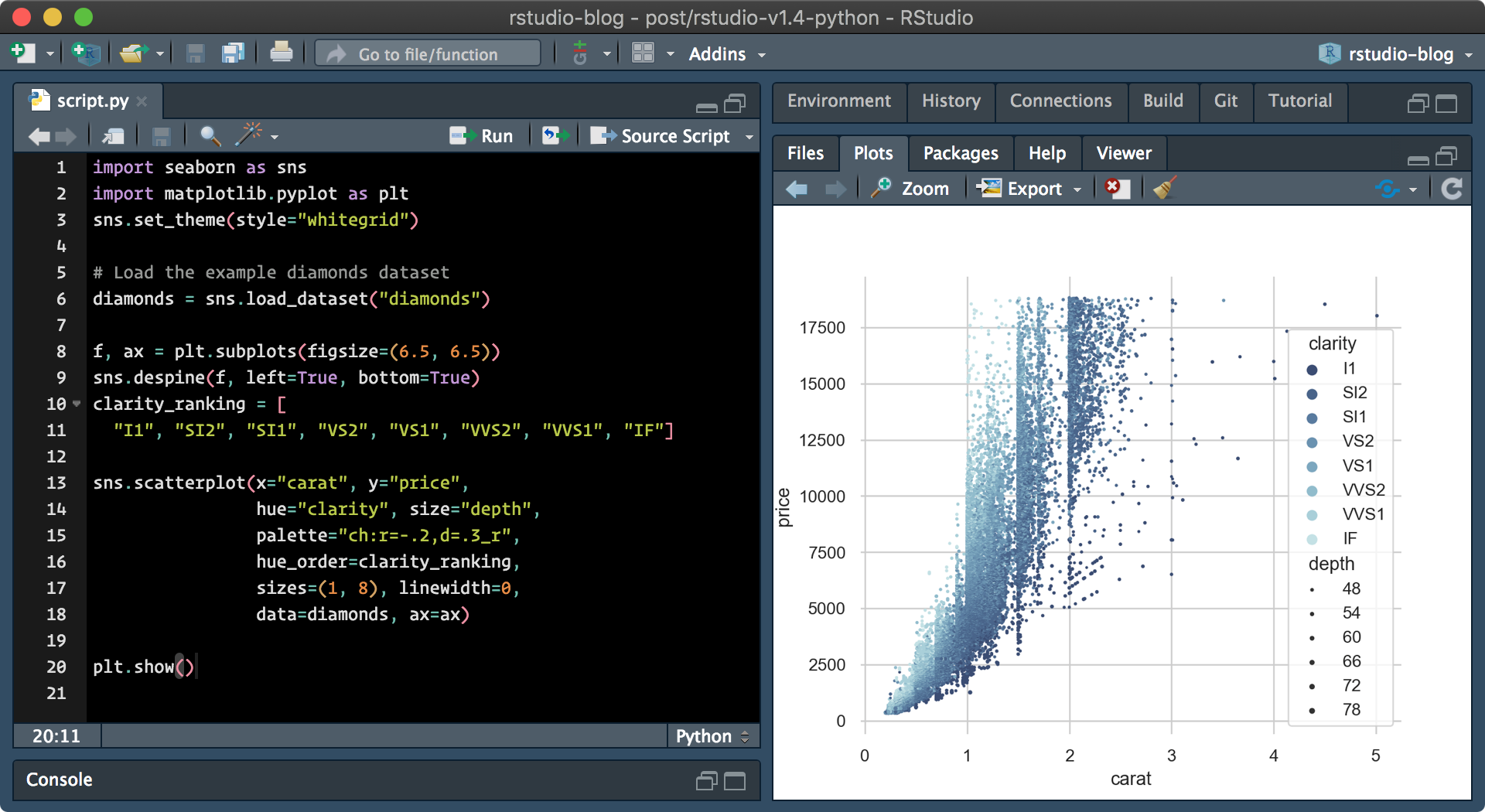
Currently, only static plots are supported, and dynamic plots may be implemented by future updates. This still means however that RStudio can render plots generated by both seaborn and matplotlib for visualizations. Additionally,matplotlib plots can now be displayed within the Plots pane when show() is called.
–
So Why Integrate?
Data scientists no longer have to choose between R or Python – especially when data science teams today have the ability to leverage both R and Python in their projects.
Under the same platform there is:
- Easier collaboration and sharing work among teams
- Less cost on a project for organization and maintenance of projects with the elimination of additional overhead
- Less effort for the maintenance, management, and scale of projects that use separate environments of different languages.
Like Anaconda, the data science ecosystem can be readily accessible by one platform.
Configuring Python in RStudio:
So to configure python with RStudio, make sure to first install a base version of Python.
Then install the reticulate package from CRAN:
install.packages("reticulate")
reticulate::py_config()
reticulate uses the Python version from your PATH which can be checked through:
Sys.which("python")) -> "C:\Users\ferdi\AppData\Local\Programs\Python\PYTHON~1\python.exe"
You can also enable virtual/conda environments:
library(reticulate)
use_virtualenv("myenv") or use_condaenv("myenv")
Now, you can run different chunks using both python and R:

–
For more resources: